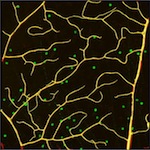
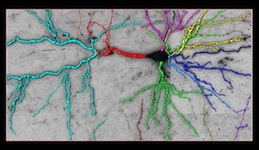
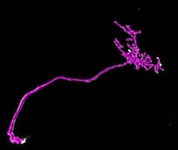
click on the images to jump to the corresponding results.
Networks of curvilinear structures are pervasive both in natural and man-made systems. They appear at all possible scales, ranging from nanometers in Electron Microscopy images of neurons and meters in aerial images of roads to petameters in dark-matter arbors binding massive galaxy clusters.
Current practical approaches for delineation require extensive manual intervention. In neuroscience research, the requirement for such editing dramatically slows down the process and makes it impossible to exploit the vast amount of data that modern microscopes can produce. As a result, there has recently been a resurgence of interest in the automated delineation problem. Yet, full automation remains elusive when the image data is noisy and the structures exhibit complex morphology.
Part of the problem comes from the fact that both greedy tracking and graph-based techniques employ hand-designed cost functions and perform heuristic optimization. As a result, they do not generalize well to different modalities and lack robustness to occlusions arising from insufficient imaging resolution and imaging artifacts such as noise, inhomogeneous contrasts, non-uniform illumination, and scene clutter.
Furthermore, these techniques rely on the assumption that curvilinear structures have a tree topology without loops. However, in practice, many interesting networks, such as those formed by the roads and blood vessels are loopy. Furthermore, even among those that really are trees, such as neuronal arbors, the imaging resolution is often so low that the branches appear to cross, thus introducing several false loops that can only be recognized as such once the whole structure has been recovered.
We introduce a novel Bayesian framework, which involves building a curvilinear network that is potentially loopy and provably very close to the global optimum of a Quadratic Mixed Integer Program (Q-MIP). We further propose an original classification-based approach to assessing the probability that a tubular path corresponds to a real curvilinear structure. This is in contrast to more traditional approaches to scoring paths by integrating pixel values along their length, which often fails to adequately penalize short-cuts and makes it difficult to compute commensurate scores for paths of different lengths.
Engin Turetken, Fethallah Benmansour, Bjoern Andres, German Gonzalez, Christian Blum, Hanspeter Pfister, Pascal Fua
Algorithm
These are the intermediate steps of our algorithm:
[su_tabs]
[su_tab title=”Image” disabled=”no” anchor=”” url=”” target=”blank” class=””]

An Aerial Image of Roads
We evaluated our approach on 3D microscopy image stacks of neurites and 2D aerial images of road networks.
[/su_tab]
[su_tab title=”Tubularity” disabled=”no” anchor=”” url=”” target=”blank” class=””]
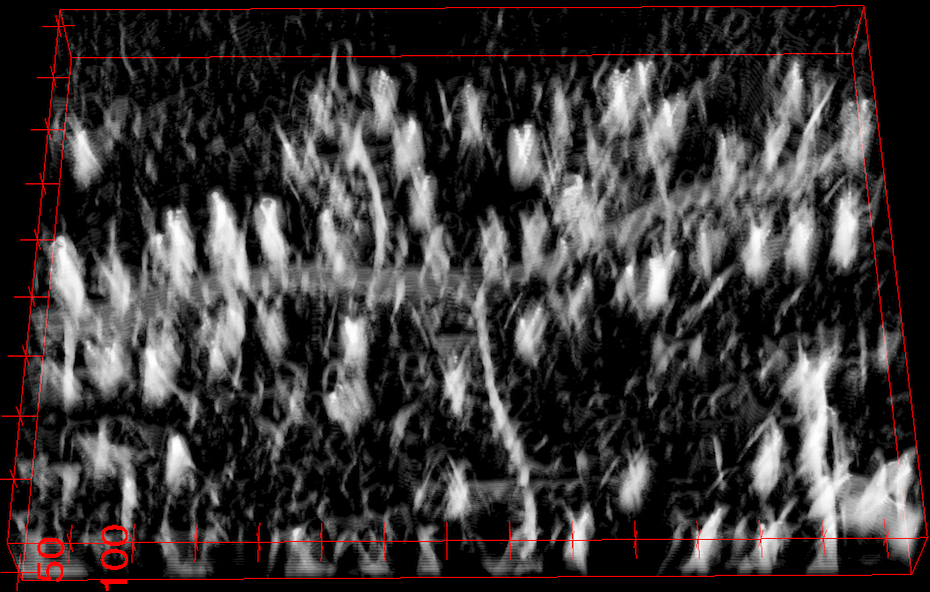
Scale-Space Tubularity Measure
We first compute a tubularity value at each image location x and radius value r, which quantifies the likelihood that there exists a tubular structure of radius r, at location x. Given an N-D image, this creates an (N+1)-D scale-space tubularity image.
[/su_tab]
[su_tab title=”Overcomplete graph” disabled=”no” anchor=”” url=”” target=”blank” class=””]
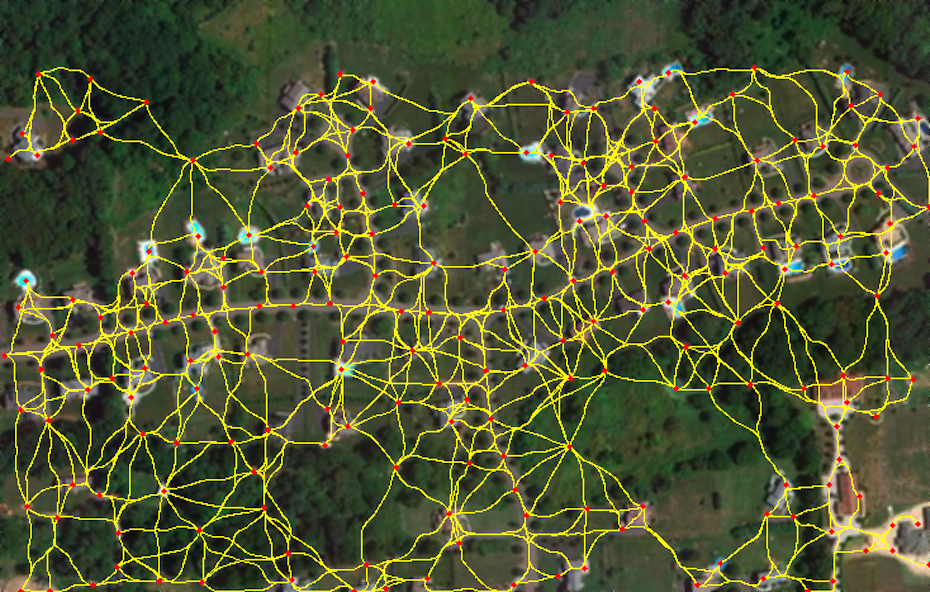
Graph over Seed Points
We select high-tubularity pixels in this image as seed points and connect them through high-tubularity paths in scale-space. This results in a directed graph.
[/su_tab]
[su_tab title=”Weighted graph” disabled=”no” anchor=”” url=”” target=”blank” class=””]
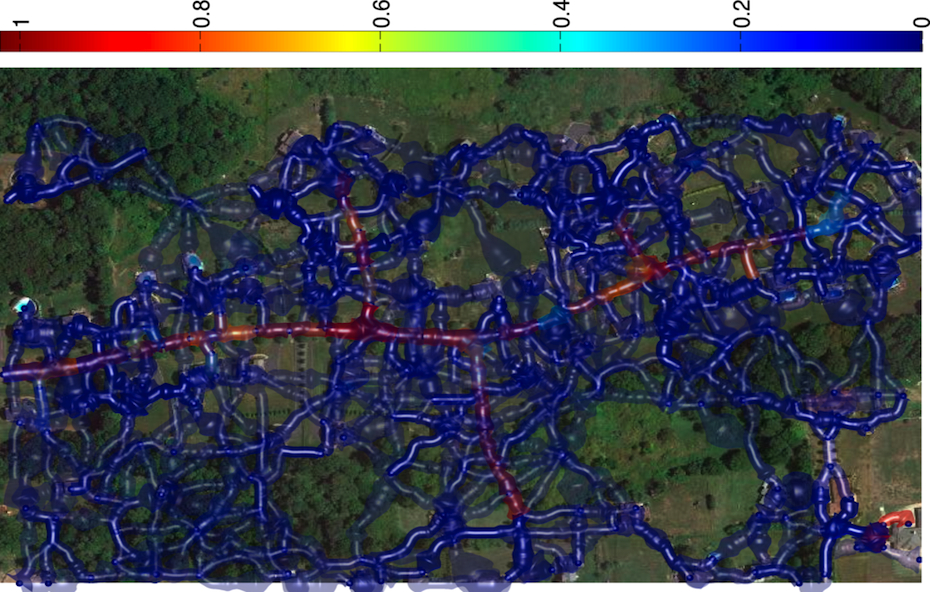
Weighted Graph Representing Path Classification Scores
Having trained a path classifier using such graphs and ground-truth trees, we assign log-likelihood ratio weights to pairs of consecutive edges of a given graph at detection time.
[/su_tab]
[su_tab title=”Weighted graph with tubularity” disabled=”no” anchor=”” url=”” target=”blank” class=””]
![]()
Weighted Graph Representing Integrated Tubularity Values
Conventional methods compute edge weights as integrating a function of the tubularity values along the paths. This results in much less informative weights.
[/su_tab]
[su_tab title=”Reconstructed tree” disabled=”no” anchor=”” url=”” target=”blank” class=””]
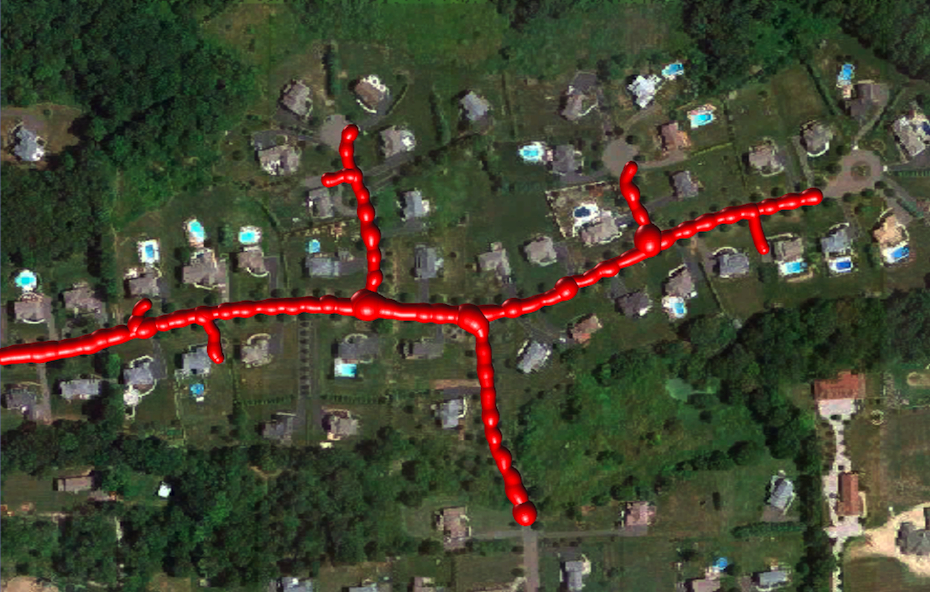
Reconstructed Tree
Finally, we compute the minimum-weight (maximum-likelihood) directed tree that spans only a subset of the edges by solving a Quadratic Mixed Integer Program.
[/su_tab]
[/su_tabs]
Results
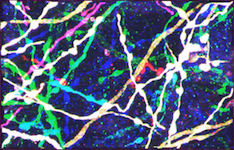
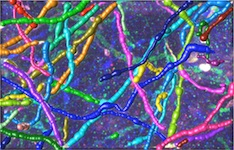
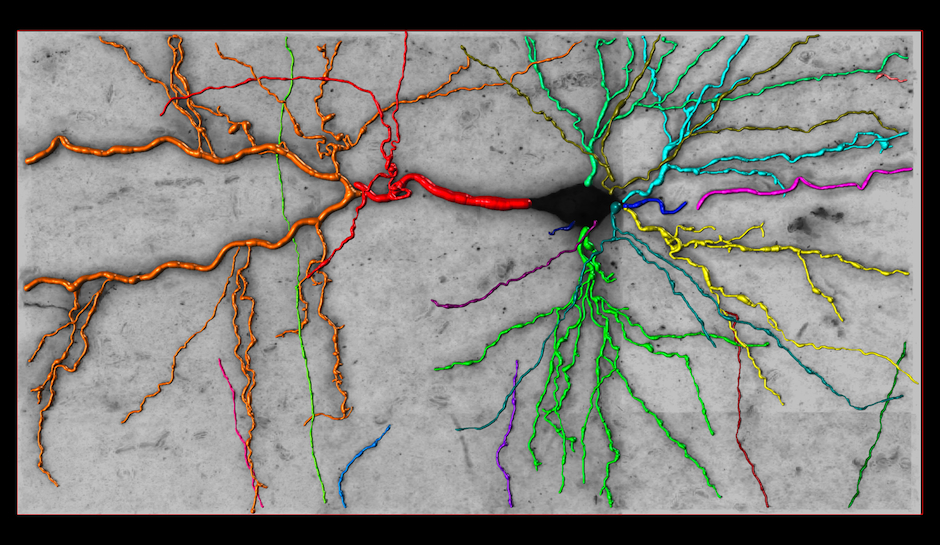
Reconstruction in a brainbow micrograph [top]
This image stack is acquired by our collaborators Luke Bogart, Takao Hensch and Jeff Lichtman from Harvard University. The neurites in this dataset are genetically engineered so that each is labeled with a distinctive color. They form tree structures without cycles in them. However, because of the low resolution, branches that are physically disjoint appear to cross, introducing several false loops. Our approach accounts for these loops and yields better network topologies. For more information about the modality, see Livet et al., 2007.
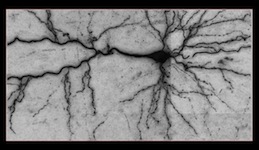
Reconstruction in a brightfield micrograph [top]
This image stack is acquired by our colleagues from EPFL’s Brain Mind Institute. The images were obtained from biocityne-dyed rat brains. The numerous artifacts produced by irregularities of the staining process and the non-Gaussian blur introduced by the microscope make their automated analysis challenging. Furthermore, many significant processes appear as faint structures, present abrupt intensity changes, or are severely blurred. Finally, similar to the brainbow stacks, dendrites and axons form tree structures, but their branches often appear to cross along the z-axis. Our algorithm eliminates both structured and unstructured noise, and produces topologically plausible trees despite branch crossings.
Embed of video is only possible from Mediaspace, SwitchTube, Vimeo or Youtube
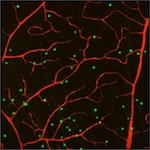
Reconstruction in confocal micrographs [top]
These two image stacks of blood vessels are acquired by our collaborators Maximilian Joesch and Markus Meister from Harvard University. Our results contain many real cycles, which are created by capillaries connecting arteries to veins. We only considered the red channel of these stacks since it is the only one used to label the blood vessels. The green blobs represent neuron nuclei.
Embed of video is only possible from Mediaspace, SwitchTube, Vimeo or Youtube
Reconstructions of the Olfactory Projection Fibers from the DIADEM Challenge [top]
These stacks are acquired from the olfactory bulb of Drosophila fly using 2-channel confocal microscopy. For additional details on the dataset, please refer to the DIADEM website.
Embed of video is only possible from Mediaspace, SwitchTube, Vimeo or Youtube
Embed of video is only possible from Mediaspace, SwitchTube, Vimeo or Youtube
Additional Comparative Results [top]
These animations provide a visual comparison of our reconstructions, manually annotated ground truths and our previous approach based on the k Minimum Spanning Tree formulation for three different modalities.
[su_tabs]
[su_tab title=”Ground Truth” disabled=”no” anchor=”” url=”” target=”blank” class=””]
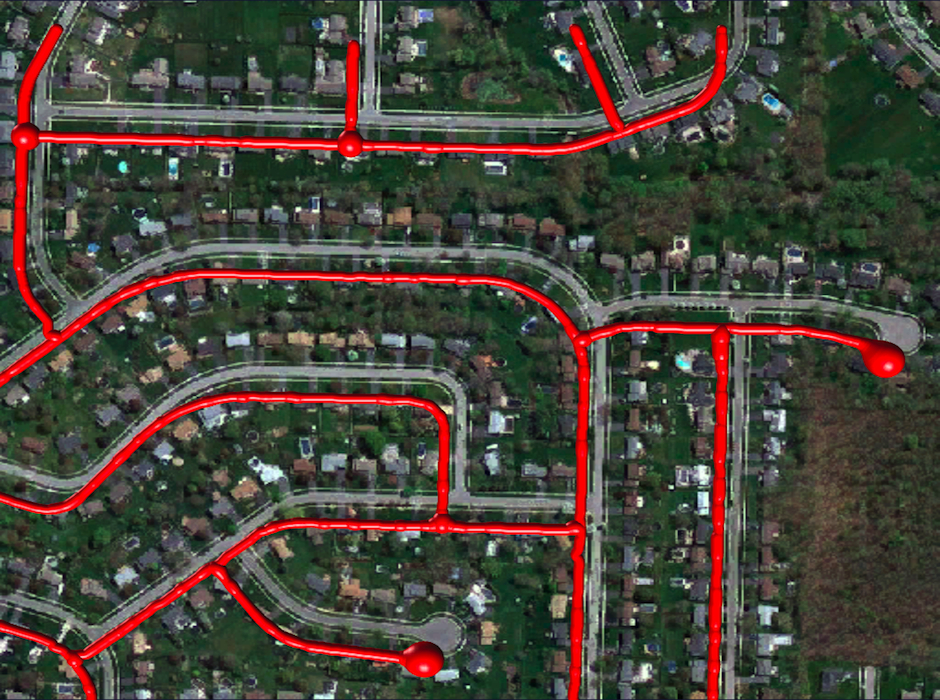
Ground Truth
The paths are drawn as consecutive spheres whose radius corresponds to the estimated width. They have been shifted from their true position to make the underlying linear structures visible.
[/su_tab]
[su_tab title=”Tubular Graph” disabled=”no” anchor=”” url=”” target=”blank” class=””]
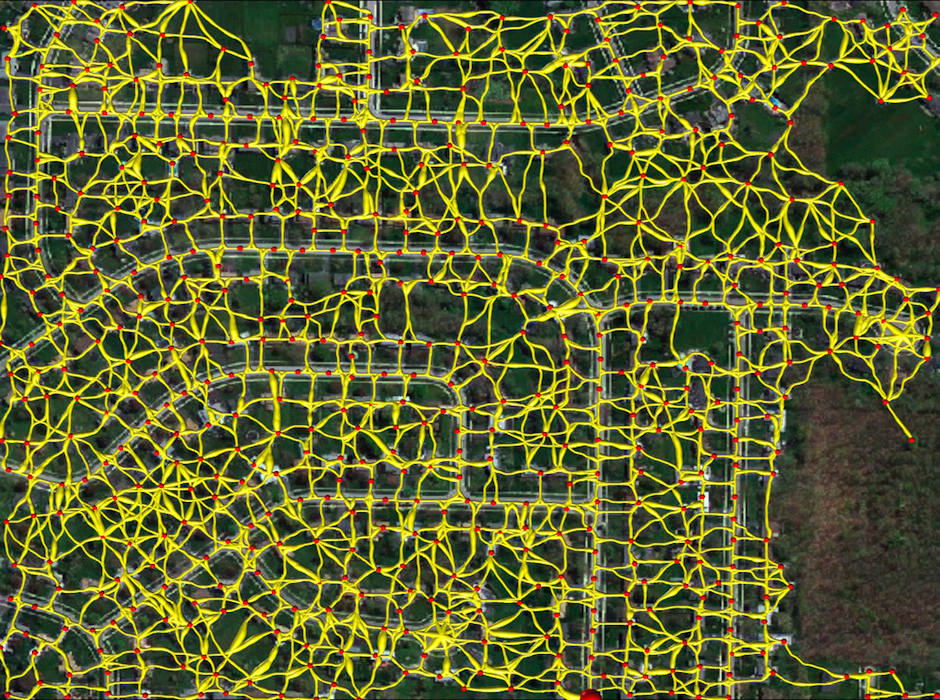
Tubular Graph
The large red sphere denotes the root vertex of the tree structure. Path widths are downscaled to allow a better visualization.
[/su_tab]
[su_tab title=”Reconstruction (k-MST)” disabled=”no” anchor=”” url=”” target=”blank” class=””]
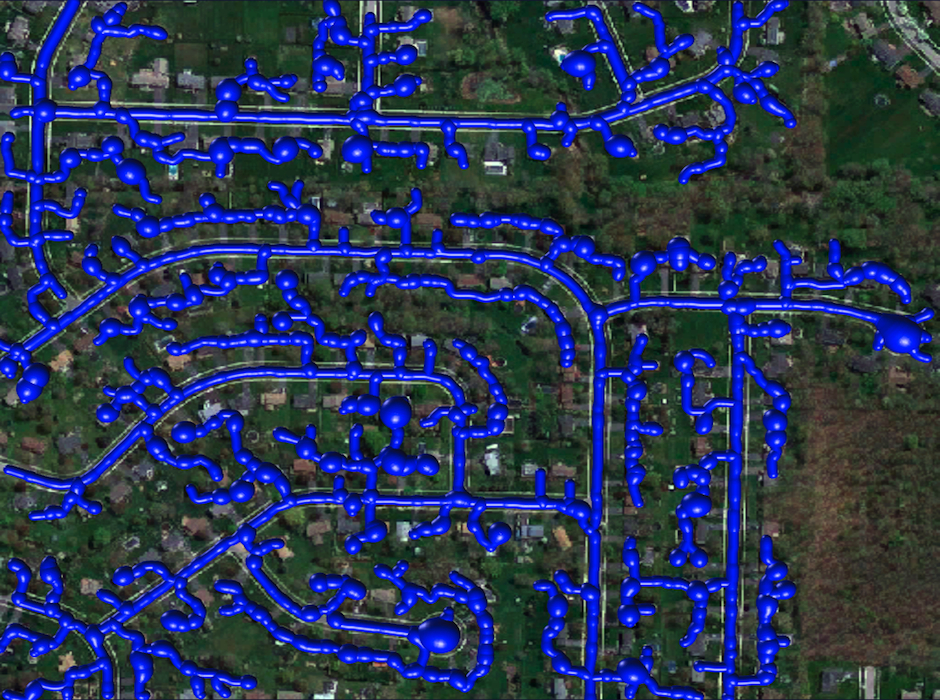
Reconstruction Obtained by the k-MST Algorithm
[/su_tab]
[su_tab title=”Reconstruction (ours)” disabled=”no” anchor=”” url=”” target=”blank” class=””]
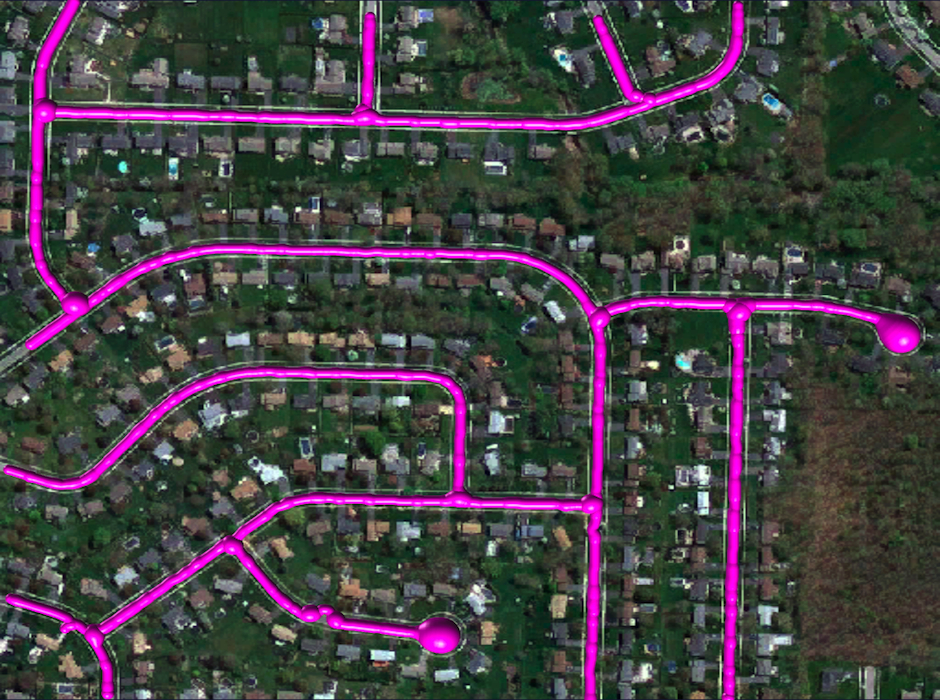
Reconstruction Obtained by Our Method
[/su_tab]
[/su_tabs]
[su_tabs]
[su_tab title=”Ground Truth” disabled=”no” anchor=”” url=”” target=”blank” class=””]
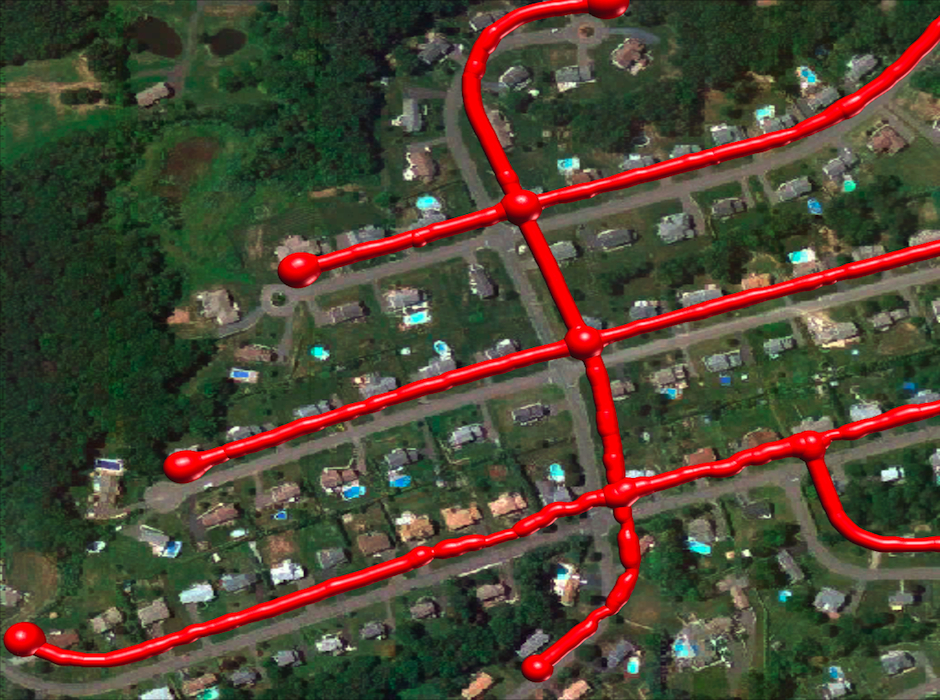
Ground Truth
[/su_tab]
[su_tab title=”Tubular Graph” disabled=”no” anchor=”” url=”” target=”blank” class=””]
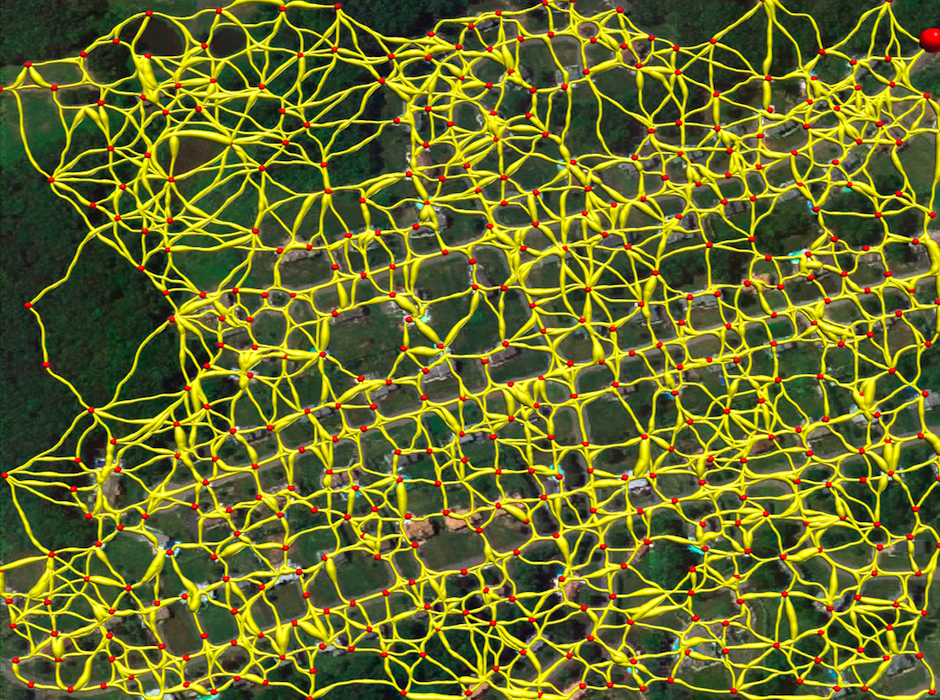
Tubular Graph
[/su_tab]
[su_tab title=”Reconstruction (k-MST)” disabled=”no” anchor=”” url=”” target=”blank” class=””]
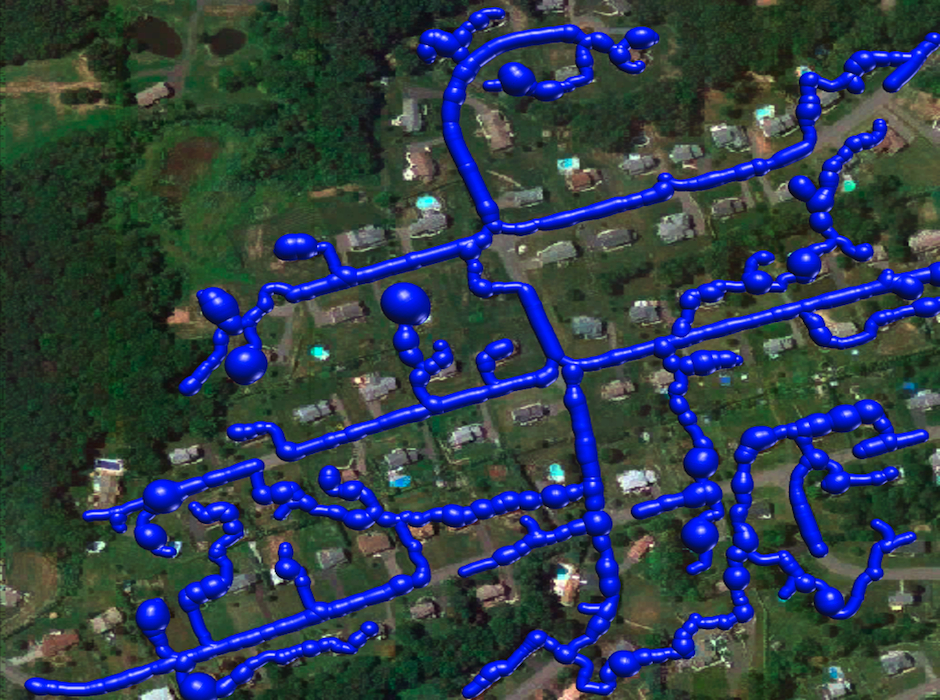
Reconstruction Obtained by the k-MST Algorithm
[/su_tab]
[su_tab title=”Reconstruction (ours)” disabled=”no” anchor=”” url=”” target=”blank” class=””]
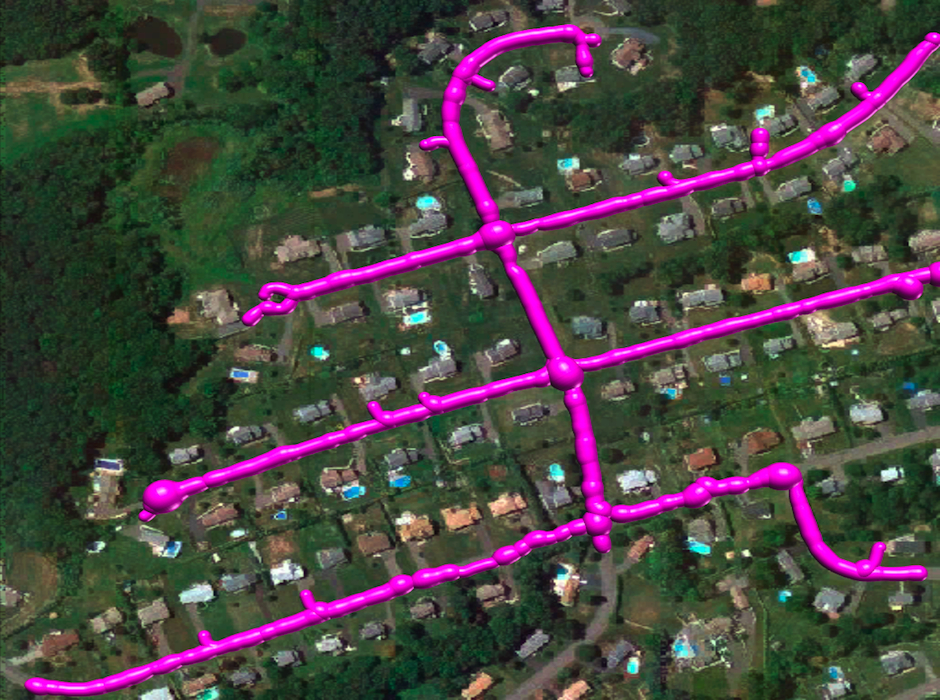
Reconstruction Obtained by Our Method
[/su_tab]
[/su_tabs]
[su_tabs]
[su_tab title=”Ground Truth” disabled=”no” anchor=”” url=”” target=”blank” class=””]
Ground Truth
[/su_tab]
[su_tab title=”Tubular Graph” disabled=”no” anchor=”” url=”” target=”blank” class=””]
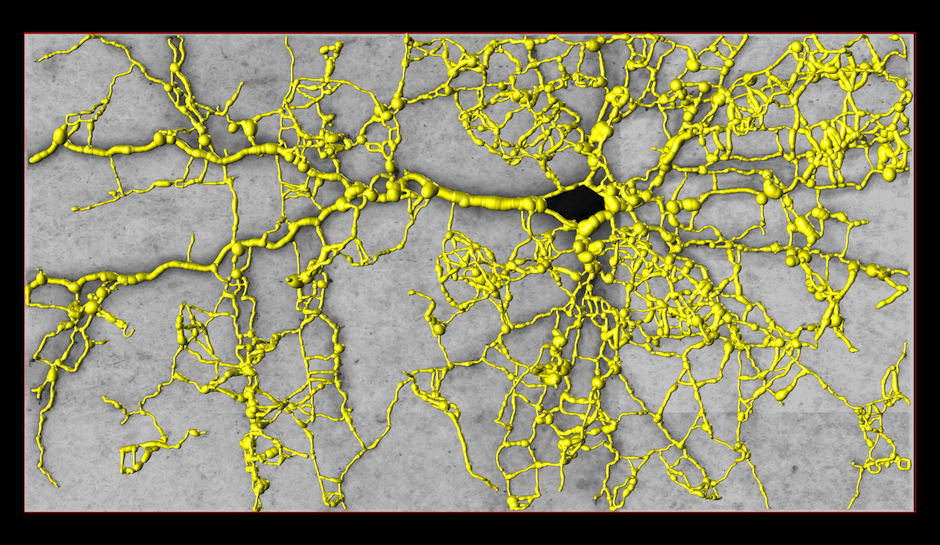
Tubular Graph
[/su_tab]
[su_tab title=”Reconstruction (k-MST)” disabled=”no” anchor=”” url=”” target=”blank” class=””]
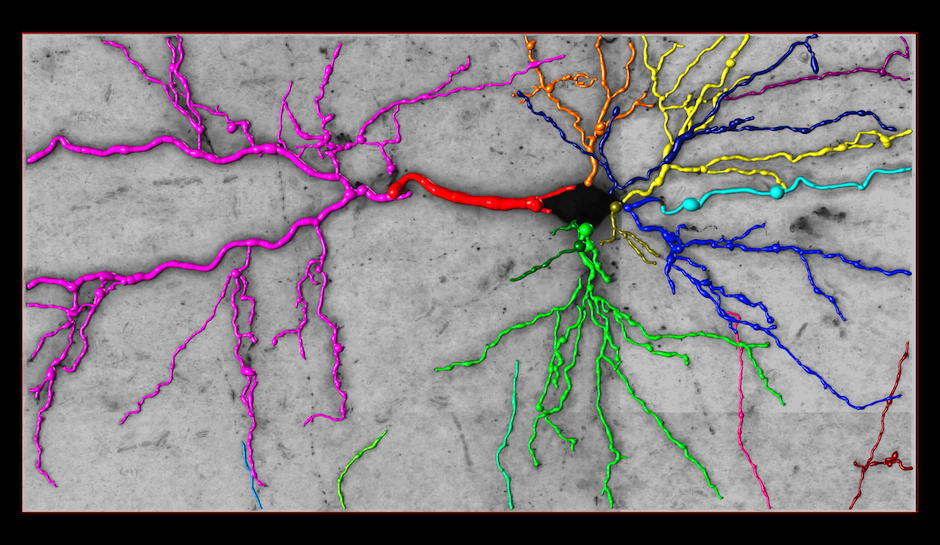
Reconstruction Obtained by the k-MST Algorithm
[/su_tab]
[su_tab title=”Reconstruction (ours)” disabled=”no” anchor=”” url=”” target=”blank” class=””]
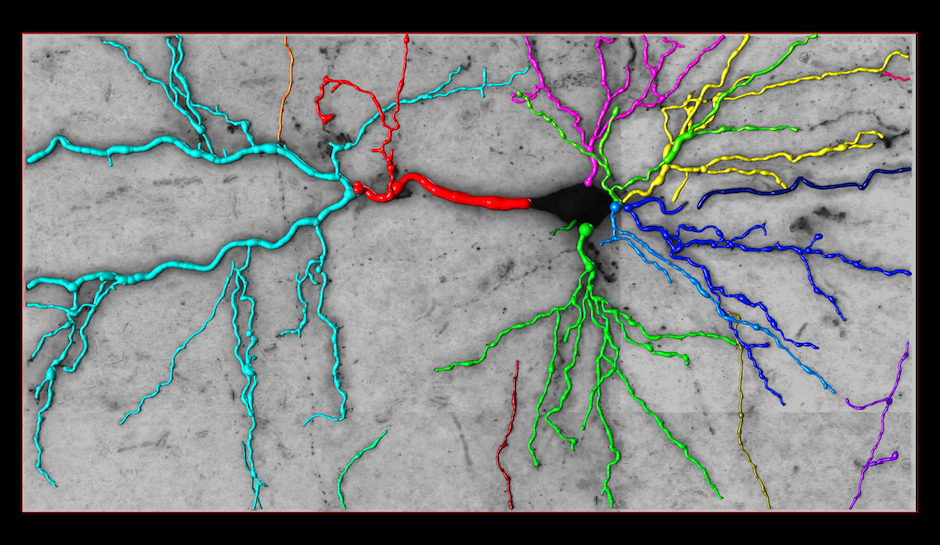
Reconstruction Obtained by Our Method
[/su_tab]
[/su_tabs]
[su_tabs]
[su_tab title=”Ground Truth” disabled=”no” anchor=”” url=”” target=”blank” class=””]
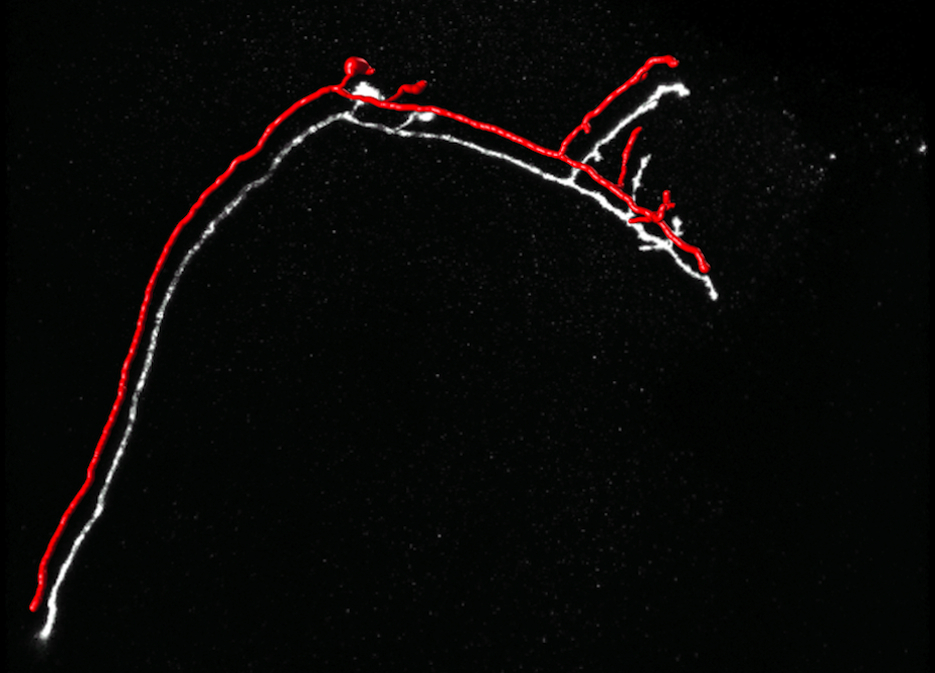
Ground Truth
[/su_tab]
[su_tab title=”Tubular Graph” disabled=”no” anchor=”” url=”” target=”blank” class=””]
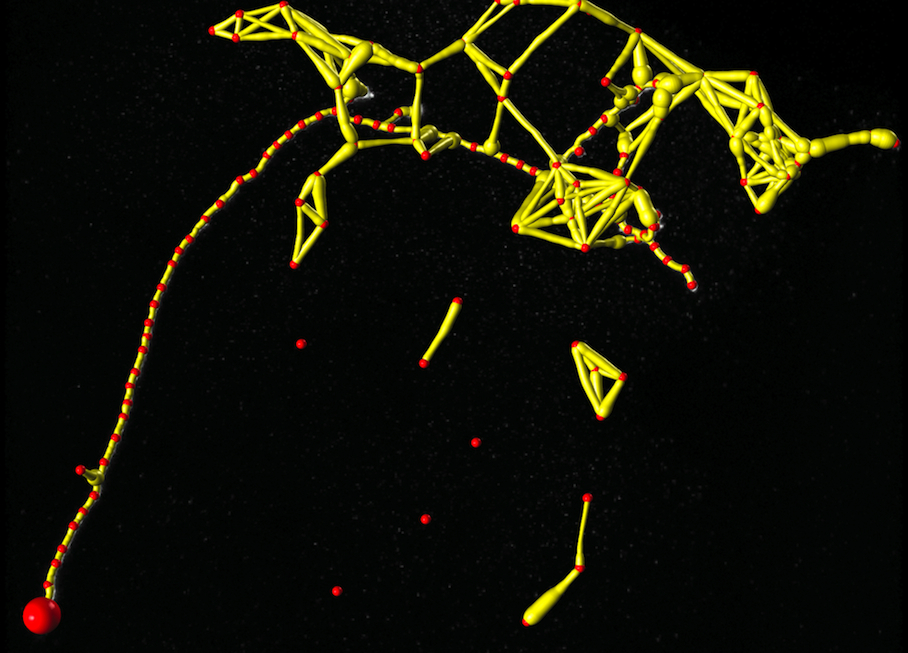
Tubular Graph
[/su_tab]
[su_tab title=”Reconstruction (k-MST)” disabled=”no” anchor=”” url=”” target=”blank” class=””]
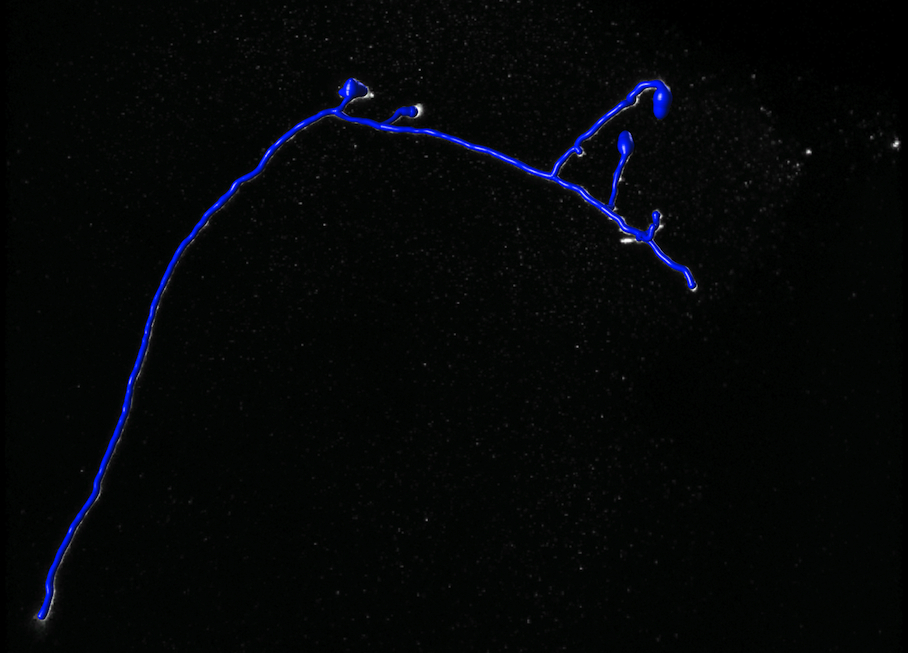
Reconstruction Obtained by the k-MST Algorithm
[/su_tab]
[su_tab title=”Reconstruction (ours)” disabled=”no” anchor=”” url=”” target=”blank” class=””]
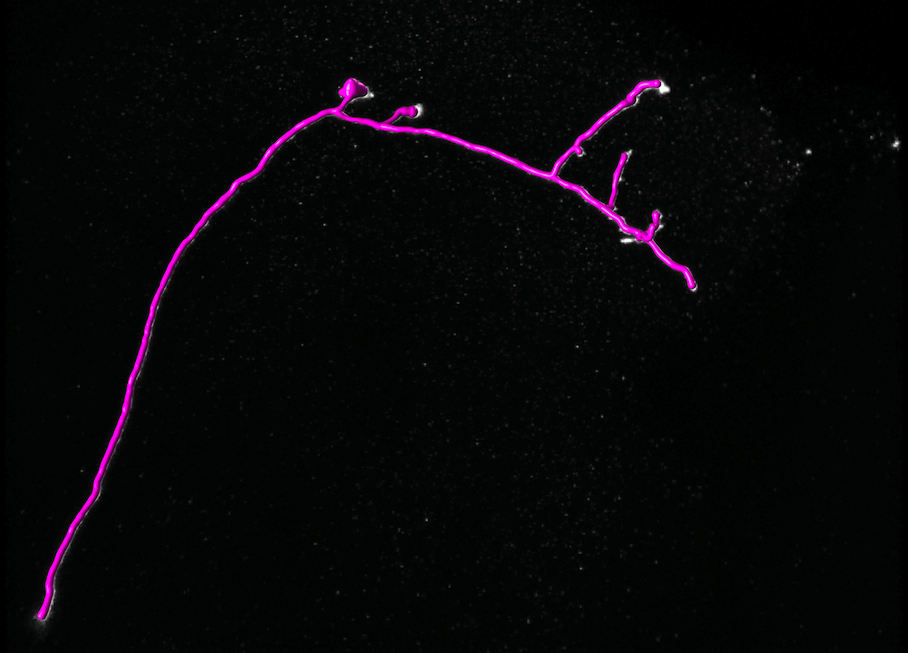
Reconstruction Obtained by Our Method
[/su_tab]
[/su_tabs]
Data
Datasets with ground truth tracings are available here.
Software
Our semi-automated tracing tool for acquiring ground-truth is available here.
References
Reconstructing Curvilinear Networks using Path Classifiers and Integer Programming
IEEE Transactions On Pattern Analysis And Machine Intelligence (PAMI). 2016. Vol. 38, num. 12, p. 2515–2530. DOI : 10.1109/Tpami.2016.2519025.Automated Reconstruction of Curvilinear Networks from 2D and 3D Imagery
Lausanne, EPFL, 2013.Multiscale Centerline Detection
IEEE Transactions on Pattern Analysis and Machine Intelligence. 2016. Vol. 38, num. 7, p. 1327–1341. DOI : 10.1109/Tpami.2015.2462363.Automated Reconstruction of Dendritic and Axonal Trees by Global Optimization with Geometric Priors
Neuroinformatics. 2011. Vol. 9, p. 279-302. DOI : 10.1007/s12021-011-9122-1.Contacts
| Engin Turetken (primary contact) | [e-mail] |
| Pascal Fua | [e-mail] |